Inverse design of 1X2 splitter
Inverse design of 1X2 splitter
Inverse Splitter using topology optimization
MEEP simulation for inverse splitter
Source Code
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
from functools import partial
from pathlib import Path
import gdsfactory as gf
import jax
import glob
import jax.numpy as jnp
import matplotlib.pyplot as plt
from PIL import Image
import numpy as np
import sax
import meep as mp
import gplugins
import gplugins.gmeep as gm
from meep import MaterialGrid, Medium, Vector3, Volume
from meep.adjoint import (
DesignRegion,
get_conic_radius_from_eta_e,
)
from autograd import numpy as npa, tensor_jacobian_product, grad
import gplugins.tidy3d as gt
from gplugins.common.config import PATH
import inspect
import meep.adjoint as mpa
from matplotlib.animation import FuncAnimation, PillowWriter
import os
1
2
Using MPI version 4.1, 1 processes
[32m2025-12-09 12:58:38.473[0m | [1mINFO [0m | [36mgplugins.gmeep[0m:[36m<module>[0m:[36m39[0m - [1mMeep '1.31.0' installed at ['/home/ramprakash/anaconda3/envs/si_photo/lib/python3.13/site-packages/meep'][0m
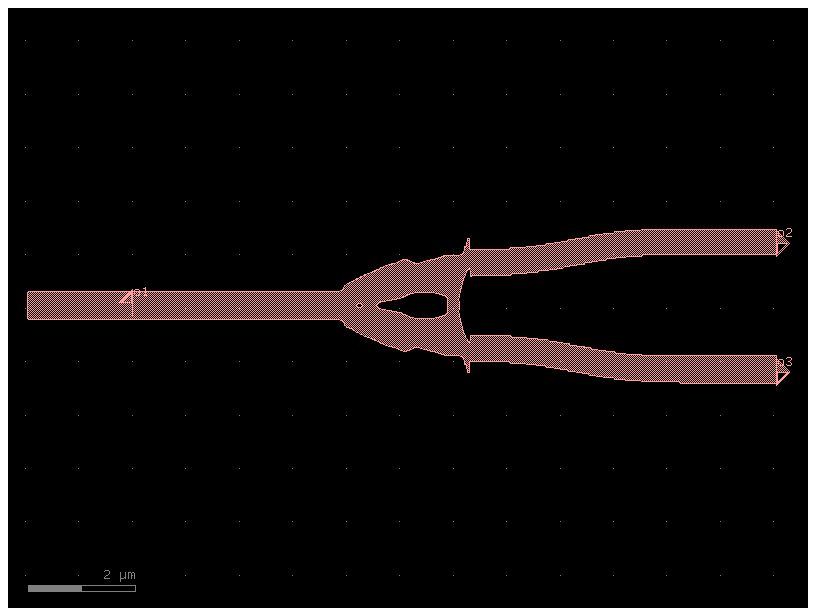
Simple ybranch from gdsfactory
Source Code
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
gf.clear_cache()
ybranch = gf.Component("ybranch")
inwg = gf.components.straight(length=5)
outwg = gf.components.straight(length=2)
splitter = gf.components.mmi1x2_with_sbend()
inwg_ref = ybranch.add_ref(inwg)
splitter_ref = ybranch.add_ref(splitter)
outwg_top_ref = ybranch.add_ref(outwg)
outwg_bot_ref = ybranch.add_ref(outwg)
# Connecting ports
inwg_ref.connect("o2", splitter_ref.ports["o1"])
outwg_top_ref.connect("o1", splitter_ref.ports["o2"])
outwg_bot_ref.connect("o1", splitter_ref.ports["o3"])
# adding port name
ybranch.add_port(name=f"o1", port=inwg_ref.ports["o1"])
ybranch.add_port(name=f"o2", port=outwg_top_ref.ports["o2"])
ybranch.add_port(name=f"o3", port=outwg_bot_ref.ports["o2"])
ybranch.auto_rename_ports()
# plotting
ybranch.draw_ports()
ybranch.plot()
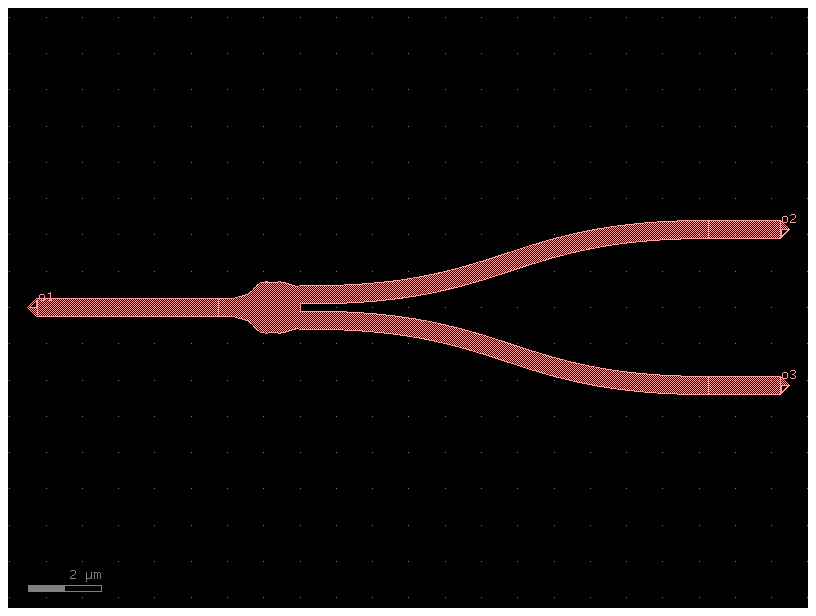
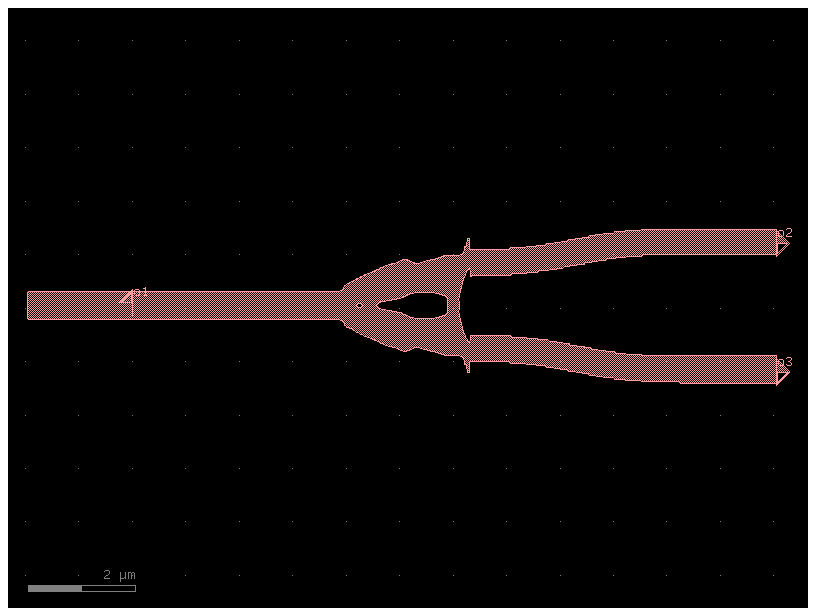
Inverse design of the splitter
Source Code
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
design_region_width = 2.5
design_region_height = 2.5
eta_e = 0.55
minimum_length = 0.1
filter_radius = get_conic_radius_from_eta_e(minimum_length, eta_e)
eta_i = 0.5
eta_d = 1 - eta_e
resolution = 20
design_region_resolution = int(5 * resolution)
Nx = int(design_region_resolution * design_region_width)
Ny = int(design_region_resolution * design_region_height)
pml_size = 1.0
waveguide_length = 1.5
waveguide_width = 0.5
Sx = 2 * pml_size + 2 * waveguide_length + design_region_width
Sy = 2 * pml_size + design_region_height + 0.5
cell_size = (Sx, Sy)
SiO2 = Medium(index=1.44)
Si = Medium(index=3.4)
Source Code
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
# Get the correct dimensions from conic_filter's mesh_grid
from meep.adjoint.filters import mesh_grid
Nx_filter, Ny_filter, _, _ = mesh_grid(get_conic_radius_from_eta_e(minimum_length, eta_e), design_region_width, design_region_height, design_region_resolution)
design_variables = MaterialGrid(Vector3(Nx_filter, Ny_filter), SiO2, Si, grid_type="U_MEAN")
design_region = DesignRegion(
design_variables,
volume=Volume(
center=Vector3(),
size=Vector3(design_region_width, design_region_height, 0),
),
)
c = gf.Component()
arm_separation = 1.0
straight1 = c << gf.components.straight(length=4, cross_section='strip')
straight1.dmove(straight1.ports["o2"].dcenter, (-design_region_width / 2.0, 0))
straight2 = c << gf.components.straight(length=2, cross_section='strip')
straight2.dmove(straight2.ports["o2"].dcenter, (design_region_width / 2.0, 0.8))
bend2 = c << gf.components.bend_s(size=(4,0.4),cross_section='strip')
bend2.connect("o1", straight2.ports["o2"])
straight3 = c << gf.components.straight(length=2, cross_section='strip')
straight3.dmove(straight3.ports["o2"].dcenter, (design_region_width / 2.0, -0.8))
bend3 = c << gf.components.bend_s(size=(4,0.4),cross_section='strip').copy()
bend3.mirror_y()
bend3.connect("o1", straight3.ports["o2"])
# bend3.dmove(straight3.ports["o1"].dcenter, (design_region_width / 2.0, -1))
c.add_port("o1", port=straight1.ports["o1"])
c.add_port("o2", port=bend2.ports["o2"])
c.add_port("o3", port=bend3.ports["o2"])
# c.draw_ports()
c.plot()
Source Code
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
def mapping(x, eta, beta):
# filter
filtered_field = mpa.conic_filter(
x,
filter_radius,
design_region_width,
design_region_height,
design_region_resolution,
)
# projection
projected_field = mpa.tanh_projection(filtered_field, beta, eta)
projected_field = (
npa.fliplr(projected_field) + projected_field
) / 2 # up-down symmetry
# interpolate to actual materials
return projected_field.flatten()
seed = 240
np.random.seed(seed)
# Get the correct dimensions from conic_filter's mesh_grid
from meep.adjoint.filters import mesh_grid
Nx_filter, Ny_filter, _, _ = mesh_grid(filter_radius, design_region_width, design_region_height, design_region_resolution)
x0 = mapping(
np.random.rand(Nx_filter * Ny_filter),
eta_i,
128,
)
def J(source, top, bottom):
"""
Objective function: Maximize total transmission to both output ports
CRITICAL FIX: Previous version divided by source, creating a ratio metric
that doesn't optimize for maximum absolute transmission.
Now we maximize the actual power transmitted.
"""
# Calculate absolute power at each output
power_top = npa.abs(top) ** 2
power_bottom = npa.abs(bottom) ** 2
# Total transmitted power (this is what we want to maximize)
total_power = power_top + power_bottom
# penalize imbalance to encourage 50/50 splitting
imbalance_penalty = 0.1 * npa.mean(npa.abs(power_top - power_bottom))
return npa.mean(total_power) - imbalance_penalty
# return npa.mean(total_power)
resolution=20
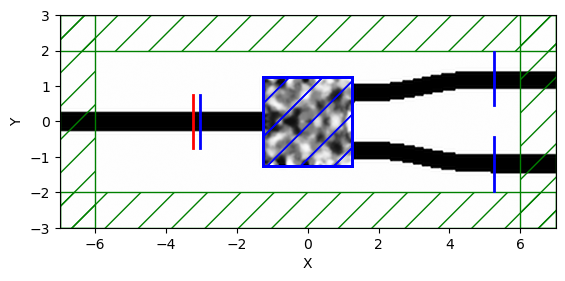
opt = gm.get_meep_adjoint_optimizer(
c,
J,
[design_region],
[design_variables],
x0,
resolution=resolution,
cell_size=(
Sx + 2 + design_region_width + 2 * pml_size,
design_region_height + 2 * pml_size + 1.5,
),
tpml=1.0,
extend_ports_length=4.0, # Extend ports to create space for sources/monitors
port_margin=0.5,
port_source_offset=-2, # Place sources just inside the extended ports
port_monitor_offset=-0, # Place monitors slightly further in
symmetries=[mp.Mirror(direction=mp.Y)],
wavelength_points=10,
)
1
2
/home/ramprakash/anaconda3/envs/si_photo/lib/python3.13/site-packages/meep/__init__.py:4446: ComplexWarning: Casting complex values to real discards the imaginary part
return _meep._get_epsilon_grid(gobj_list, mlist, _default_material, _ensure_periodicity, gv, cell_size, cell_center, nx, xtics, ny, ytics, nz, ztics, grid_vals, frequency)
Source Code
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
evaluation_history = []
cur_iter = [0]
# Create directory for saving frames
os.makedirs('optimization_frames_2', exist_ok=True)
def f(v, gradient, cur_beta):
print(f"Current iteration: {cur_iter[0] + 1}")
f0, dJ_du = opt([mapping(v, eta_i, cur_beta)])
# Save the design plot as an image instead of showing it
fig, ax = plt.subplots(figsize=(10, 8))
opt.plot2D(
False,
ax=ax,
plot_sources_flag=True,
plot_monitors_flag=True,
plot_boundaries_flag=False,
)
ax.set_title(f'Iteration {cur_iter[0] + 1}, FOM: {np.max(np.real(f0)):.4f}')
plt.savefig(f'optimization_frames_2/design_{cur_iter[0]:04d}.png', dpi=100, bbox_inches='tight')
plt.close(fig)
if gradient.size > 0:
gradient[:] = tensor_jacobian_product(mapping, 0)(
v, eta_i, cur_beta, np.sum(dJ_du, axis=1)
)
evaluation_history.append(np.max(np.real(f0)))
cur_iter[0] = cur_iter[0] + 1
return np.real(f0)
Source Code
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
# Define spatial arrays used to generate bit masks
x_g = np.linspace(-design_region_width / 2, design_region_width / 2, Nx_filter)
y_g = np.linspace(-design_region_height / 2, design_region_height / 2, Ny_filter)
X_g, Y_g = np.meshgrid(x_g, y_g, sparse=True, indexing="ij")
# IMPORTANT: Check that arm_separation matches the actual output positions
# Input at y=0, outputs at y=±1
arm_separation = 1.0
# Define the core mask - these waveguides should be FIXED to silicon
# Input waveguide at center (y=0)
left_wg_mask = (X_g == -design_region_width / 2) & (np.abs(Y_g) <= waveguide_width / 2)
# Output waveguides at y=+1 and y=-1
# Top output: centered at y=+arm_separation
top_right_wg_mask = (X_g == design_region_width / 2) & (
np.abs(Y_g - arm_separation) <= waveguide_width / 2
)
# Bottom output: centered at y=-arm_separation
bottom_right_wg_mask = (X_g == design_region_width / 2) & (
np.abs(Y_g + arm_separation) <= waveguide_width / 2
)
Si_mask = left_wg_mask | top_right_wg_mask | bottom_right_wg_mask
# Define the cladding mask - these should be FIXED to SiO2
border_mask = (
(X_g == -design_region_width / 2)
| (X_g == design_region_width / 2)
| (Y_g == -design_region_height / 2)
| (Y_g == design_region_height / 2)
)
SiO2_mask = border_mask.copy()
SiO2_mask[Si_mask] = False
# Debug: Print mask statistics
print(f"Design region: {design_region_width} x {design_region_height} um")
print(f"Grid size: {Nx_filter} x {Ny_filter}")
print(f"Si pixels (fixed): {np.sum(Si_mask)} ({100*np.sum(Si_mask)/Si_mask.size:.2f}%)")
print(f"SiO2 pixels (fixed): {np.sum(SiO2_mask)} ({100*np.sum(SiO2_mask)/SiO2_mask.size:.2f}%)")
print(f"Free pixels: {np.sum(~Si_mask & ~SiO2_mask)} ({100*np.sum(~Si_mask & ~SiO2_mask)/Si_mask.size:.2f}%)")
1
2
3
4
5
Design region: 2.5 x 2.5 um
Grid size: 251 x 251
Si pixels (fixed): 153 (0.24%)
SiO2 pixels (fixed): 847 (1.34%)
Free pixels: 62001 (98.41%)
Source Code
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
n = Nx_filter * Ny_filter # number of parameters
# Initial guess
x = np.ones((n,)) * 0.5
x[Si_mask.flatten()] = 1 # set the edges of waveguides to silicon
x[SiO2_mask.flatten()] = 0 # set the other edges to SiO2
# lower and upper bounds
lb = np.zeros((Nx_filter * Ny_filter,))
lb[Si_mask.flatten()] = 1
ub = np.ones((Nx_filter * Ny_filter,))
ub[SiO2_mask.flatten()] = 0
# IMPROVED OPTIMIZATION PARAMETERS
# Start with lower beta for better exploration
cur_beta = 4 # Changed from 4 to 2 (gentler start)
beta_scale = 2
num_betas = 6 # Changed from 7 to 8 (more gradual increase)
update_factor = 12 # Changed from 12 to 30 (MORE ITERATIONS per beta!)
# Total iterations: 8 * 30 = 240 (was only 84 before)
run_optimization = True
if run_optimization:
print(f"Starting optimization with {num_betas * update_factor} total iterations")
print(f"Beta schedule: {cur_beta} -> {cur_beta * (beta_scale ** (num_betas - 1))}")
for iters in range(num_betas):
print(f"\n{'='*70}")
print(f"Beta iteration {iters+1}/{num_betas}, current beta: {cur_beta}")
print(f"{'='*70}")
if iters != num_betas - 1:
x[:] = gm.run_meep_adjoint_optimizer(
n,
lambda a, g: f(a, g, cur_beta),
x,
lower_bound=lb,
upper_bound=ub,
maxeval=update_factor,
)
else:
optimized_component = gm.run_meep_adjoint_optimizer(
n,
lambda a, g: f(a, g, cur_beta),
x,
lower_bound=lb,
upper_bound=ub,
maxeval=update_factor,
get_optimized_component=True,
opt=opt,
threshold_offset_from_max=0.09,
)
cur_beta = cur_beta * beta_scale
optimized_component.plot()
# Calculate final FOM
final_fom = np.array(evaluation_history[-1])
final_transmission = np.mean(final_fom)
final_figure_of_merit_dB = 10 * np.log10(0.5 * final_transmission)
print(f"\n{'='*70}")
print(f"OPTIMIZATION COMPLETE")
print(f"{'='*70}")
print(f"Final transmission: {final_transmission:.4f} ({final_transmission*100:.2f}%)")
print(f"Expected insertion loss: ~{-10*np.log10(final_transmission):.2f} dB")
print(f"Final FOM (per port): {final_figure_of_merit_dB:.2f} dB")
print(f"Total iterations: {len(evaluation_history)}")
print(f"{'='*70}")
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
118
119
120
121
122
123
124
125
126
127
128
129
130
131
132
133
134
135
136
137
138
139
140
141
142
143
144
145
146
147
148
149
150
151
152
153
154
155
156
157
158
159
160
161
162
163
164
165
166
167
168
169
170
171
172
173
174
175
176
177
178
179
180
181
182
183
184
185
186
187
188
189
190
191
192
193
194
195
196
197
198
199
200
201
202
203
204
205
206
207
208
209
210
211
212
213
214
215
216
217
218
219
220
221
222
223
224
225
226
227
228
229
230
231
232
233
234
235
236
237
238
239
240
241
242
243
244
245
246
247
248
249
250
251
252
253
254
255
256
257
258
259
260
261
262
263
264
265
266
267
268
269
270
271
272
273
274
275
276
277
278
279
280
281
282
283
284
285
286
287
288
289
290
291
292
293
294
295
296
297
298
299
300
301
302
303
304
305
306
307
308
309
310
311
312
313
314
315
316
317
318
319
320
321
322
323
324
325
326
327
328
329
330
331
332
333
334
335
336
337
338
339
340
341
342
343
344
345
Starting optimization with 72 total iterations
Beta schedule: 4 -> 128
======================================================================
Beta iteration 1/6, current beta: 4
======================================================================
Current iteration: 1
Starting forward run...
/home/ramprakash/anaconda3/envs/si_photo/lib/python3.13/site-packages/meep/__init__.py:4440: ComplexWarning: Casting complex values to real discards the imaginary part
return _meep.create_structure(cell_size, dft_data_list_, pml_1d_vols_, pml_2d_vols_, pml_3d_vols_, absorber_vols_, gv, br, sym, num_chunks, Courant, use_anisotropic_averaging, tol, maxeval, gobj_list, center, _ensure_periodicity, _default_material, alist, extra_materials, split_chunks_evenly, set_materials, existing_s, output_chunk_costs, my_bp)
/home/ramprakash/anaconda3/envs/si_photo/lib/python3.13/site-packages/meep/__init__.py:4443: ComplexWarning: Casting complex values to real discards the imaginary part
return _meep._set_materials(s, cell_size, gv, use_anisotropic_averaging, tol, maxeval, gobj_list, center, _ensure_periodicity, _default_material, alist, extra_materials, split_chunks_evenly, set_materials, existing_geps, output_chunk_costs, my_bp)
Starting adjoint run...
/home/ramprakash/anaconda3/envs/si_photo/lib/python3.13/site-packages/autograd/tracer.py:16: UserWarning: Output seems independent of input.
warnings.warn("Output seems independent of input.")
Calculating gradient...
/home/ramprakash/anaconda3/envs/si_photo/lib/python3.13/site-packages/meep/visualization.py:971: UserWarning: plot_sources_flag is deprecated. Use show_sources instead.
warnings.warn("plot_sources_flag is deprecated. " "Use show_sources instead.")
/home/ramprakash/anaconda3/envs/si_photo/lib/python3.13/site-packages/meep/visualization.py:976: UserWarning: plot_monitors_flag is deprecated. Use show_monitors instead.
warnings.warn("plot_monitors_flag is deprecated. Use show_monitors " "instead.")
Current iteration: 2
Starting forward run...
Starting adjoint run...
Calculating gradient...
Current iteration: 3
Starting forward run...
Starting adjoint run...
Calculating gradient...
Current iteration: 4
Starting forward run...
Starting adjoint run...
Calculating gradient...
Current iteration: 5
Starting forward run...
Starting adjoint run...
Calculating gradient...
Current iteration: 6
Starting forward run...
Starting adjoint run...
Calculating gradient...
Current iteration: 7
Starting forward run...
Starting adjoint run...
Calculating gradient...
Current iteration: 8
Starting forward run...
Starting adjoint run...
Calculating gradient...
Current iteration: 9
Starting forward run...
Starting adjoint run...
Calculating gradient...
Current iteration: 10
Starting forward run...
Starting adjoint run...
Calculating gradient...
Current iteration: 11
Starting forward run...
Starting adjoint run...
Calculating gradient...
Current iteration: 12
Starting forward run...
Starting adjoint run...
Calculating gradient...
======================================================================
Beta iteration 2/6, current beta: 8
======================================================================
Current iteration: 13
Starting forward run...
Starting adjoint run...
Calculating gradient...
Current iteration: 14
Starting forward run...
Starting adjoint run...
Calculating gradient...
Current iteration: 15
Starting forward run...
Starting adjoint run...
Calculating gradient...
Current iteration: 16
Starting forward run...
Starting adjoint run...
Calculating gradient...
Current iteration: 17
Starting forward run...
Starting adjoint run...
Calculating gradient...
Current iteration: 18
Starting forward run...
Starting adjoint run...
Calculating gradient...
Current iteration: 19
Starting forward run...
Starting adjoint run...
Calculating gradient...
Current iteration: 20
Starting forward run...
Starting adjoint run...
Calculating gradient...
Current iteration: 21
Starting forward run...
Starting adjoint run...
Calculating gradient...
Current iteration: 22
Starting forward run...
Starting adjoint run...
Calculating gradient...
Current iteration: 23
Starting forward run...
Starting adjoint run...
Calculating gradient...
Current iteration: 24
Starting forward run...
Starting adjoint run...
Calculating gradient...
======================================================================
Beta iteration 3/6, current beta: 16
======================================================================
Current iteration: 25
Starting forward run...
Starting adjoint run...
Calculating gradient...
Current iteration: 26
Starting forward run...
Starting adjoint run...
Calculating gradient...
Current iteration: 27
Starting forward run...
Starting adjoint run...
Calculating gradient...
Current iteration: 28
Starting forward run...
Starting adjoint run...
Calculating gradient...
Current iteration: 29
Starting forward run...
Starting adjoint run...
Calculating gradient...
Current iteration: 30
Starting forward run...
Starting adjoint run...
Calculating gradient...
Current iteration: 31
Starting forward run...
Starting adjoint run...
Calculating gradient...
Current iteration: 32
Starting forward run...
Starting adjoint run...
Calculating gradient...
Current iteration: 33
Starting forward run...
Starting adjoint run...
Calculating gradient...
Current iteration: 34
Starting forward run...
Starting adjoint run...
Calculating gradient...
Current iteration: 35
Starting forward run...
Starting adjoint run...
Calculating gradient...
Current iteration: 36
Starting forward run...
Starting adjoint run...
Calculating gradient...
======================================================================
Beta iteration 4/6, current beta: 32
======================================================================
Current iteration: 37
Starting forward run...
Starting adjoint run...
Calculating gradient...
Current iteration: 38
Starting forward run...
Starting adjoint run...
Calculating gradient...
Current iteration: 39
Starting forward run...
Starting adjoint run...
Calculating gradient...
Current iteration: 40
Starting forward run...
Starting adjoint run...
Calculating gradient...
Current iteration: 41
Starting forward run...
Starting adjoint run...
Calculating gradient...
Current iteration: 42
Starting forward run...
Starting adjoint run...
Calculating gradient...
Current iteration: 43
Starting forward run...
Starting adjoint run...
Calculating gradient...
Current iteration: 44
Starting forward run...
Starting adjoint run...
Calculating gradient...
Current iteration: 45
Starting forward run...
Starting adjoint run...
Calculating gradient...
Current iteration: 46
Starting forward run...
Starting adjoint run...
Calculating gradient...
Current iteration: 47
Starting forward run...
Starting adjoint run...
Calculating gradient...
Current iteration: 48
Starting forward run...
Starting adjoint run...
Calculating gradient...
======================================================================
Beta iteration 5/6, current beta: 64
======================================================================
Current iteration: 49
Starting forward run...
Starting adjoint run...
Calculating gradient...
Current iteration: 50
Starting forward run...
Starting adjoint run...
Calculating gradient...
Current iteration: 51
Starting forward run...
Starting adjoint run...
Calculating gradient...
Current iteration: 52
Starting forward run...
Starting adjoint run...
Calculating gradient...
Current iteration: 53
Starting forward run...
Starting adjoint run...
Calculating gradient...
Current iteration: 54
Starting forward run...
Starting adjoint run...
Calculating gradient...
Current iteration: 55
Starting forward run...
Starting adjoint run...
Calculating gradient...
Current iteration: 56
Starting forward run...
Starting adjoint run...
Calculating gradient...
Current iteration: 57
Starting forward run...
Starting adjoint run...
Calculating gradient...
Current iteration: 58
Starting forward run...
Starting adjoint run...
Calculating gradient...
Current iteration: 59
Starting forward run...
Starting adjoint run...
Calculating gradient...
Current iteration: 60
Starting forward run...
Starting adjoint run...
Calculating gradient...
======================================================================
Beta iteration 6/6, current beta: 128
======================================================================
Current iteration: 61
Starting forward run...
Starting adjoint run...
Calculating gradient...
Current iteration: 62
Starting forward run...
Starting adjoint run...
Calculating gradient...
Current iteration: 63
Starting forward run...
Starting adjoint run...
Calculating gradient...
Current iteration: 64
Starting forward run...
Starting adjoint run...
Calculating gradient...
Current iteration: 65
Starting forward run...
Starting adjoint run...
Calculating gradient...
Current iteration: 66
Starting forward run...
Starting adjoint run...
Calculating gradient...
Current iteration: 67
Starting forward run...
Starting adjoint run...
Calculating gradient...
Current iteration: 68
Starting forward run...
Starting adjoint run...
Calculating gradient...
Current iteration: 69
Starting forward run...
Starting adjoint run...
Calculating gradient...
Current iteration: 70
Starting forward run...
Starting adjoint run...
Calculating gradient...
Current iteration: 71
Starting forward run...
Starting adjoint run...
Calculating gradient...
Current iteration: 72
Starting forward run...
Starting adjoint run...
Calculating gradient...
======================================================================
OPTIMIZATION COMPLETE
======================================================================
Final transmission: 46.0168 (4601.68%)
Expected insertion loss: ~-16.63 dB
Final FOM (per port): 13.62 dB
Total iterations: 72
======================================================================
Source Code
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
opt_device = gm.meep_adjoint_optimization.get_component_from_sim(opt.sim)
# Manually add ports at the waveguide ends
# Adjust these coordinates to match your actual waveguide positions
opt_device.add_port(
name="o1",
center=(2, 3.1), # Input port - adjust x, y as needed
width=0.5,
orientation=180, cross_section='strip'
)
opt_device.add_port(
name="o2",
center=(14.05, 4.21), # Top output port - adjust x, y as needed
width=0.5,
orientation=0, cross_section='strip'
)
opt_device.add_port(
name="o3",
center=(14.05, 1.8), # Bottom output port - adjust x, y as needed
width=0.5,
orientation=0, cross_section='strip'
)
opt_device.draw_ports()
opt_device.plot()
opt_device.show()
opt_device.write_gds("opt_device_with_ports_new_2.gds")
1
2
3
4
5
6
7
[32m2025-12-08 23:23:25.345[0m | [33m[1mWARNING [0m | [36mkfactory.kcell[0m:[36mshow[0m:[36m4100[0m - [33m[1mklive didn't send data, closing[0m
PosixPath('opt_device_with_ports_new_2.gds')
Create Animation Videos
Create animated GIFs/videos showing the optimization progress
Source Code
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
# Create combined animation showing both design and FOM
import matplotlib.gridspec as gridspec
from IPython.display import Image as IPImage
frame_files = sorted(glob.glob('optimization_frames_2/design_*.png'))
if len(frame_files) > 0 and len(evaluation_history) > 0:
fig = plt.figure(figsize=(18, 8))
gs = gridspec.GridSpec(1, 2, width_ratios=[1.5, 1])
ax1 = plt.subplot(gs[0]) # Design
ax2 = plt.subplot(gs[1]) # FOM plot
ax1.axis('off')
# Load first image
img = Image.open(frame_files[0])
im = ax1.imshow(img)
line, = ax2.plot([], [], 'b-o', linewidth=2, markersize=6)
ax2.set_xlabel('Iteration', fontsize=12)
ax2.set_ylabel('Figure of Merit', fontsize=12)
ax2.set_title('Optimization Progress', fontsize=14)
ax2.grid(True, alpha=0.3)
ax2.set_xlim(0, len(evaluation_history) + 1)
y_min = min(evaluation_history) * 0.95
y_max = max(evaluation_history) * 1.05
ax2.set_ylim(y_min, y_max)
def update_combined(frame_idx):
# Update design image
img = Image.open(frame_files[frame_idx])
im.set_data(img)
# Update FOM plot
x_data = range(1, frame_idx + 2)
y_data = evaluation_history[:frame_idx + 1]
line.set_data(x_data, y_data)
return [im, line]
anim_combined = FuncAnimation(fig, update_combined, frames=len(frame_files),
interval=200, blit=True, repeat=True)
# Save as GIF
writer = PillowWriter(fps=5)
anim_combined.save('optimization_combined_3.gif', writer=writer, dpi=100)
plt.close()
print("Combined animation saved as 'optimization_combined.gif'")
# Display the animation
display(IPImage(filename='optimization_combined_3.gif'))
else:
print("Missing data for combined animation.")
1
2
3
4
5
Combined animation saved as 'optimization_combined.gif'
<IPython.core.display.Image object>
TODO calcualte the sparammeter for the inverse design and the generic design and compare them.
Next, build a compact model for the inverse design and then make an mzi
Source Code
1
2
opt_device = gf.import_gds(gdspath="opt_device_with_ports_new_2.gds")
opt_device.plot()
1
2
3
[32m2025-12-08 23:23:30.943[0m | [31m[1mERROR [0m | [36mkfactory.kcell[0m:[36mname[0m:[36m687[0m - [31m[1mName conflict in kfactory.kcell::name at line 687
Renaming Unnamed_32 (cell_index=32) to Unnamed_31 would cause it to be named the same as:
- Unnamed_31 (cell_index=31), function_name=None, basename=None[0m
MEEP FDTD simualtion to do sparameter extraction
Source Code
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
wavelengths = np.linspace(1.5, 1.6, 50)
lda_c = wavelengths[wavelengths.size // 2]
pdk = gf.get_active_pdk()
layer_stack = pdk.get_layer_stack()
core = layer_stack.layers["core"]
clad = layer_stack.layers["clad"]
box = layer_stack.layers["box"]
layer_stack.layers.pop("substrate", None)
print(
f"""Stack:
- {clad.material} clad with {clad.thickness}µm
- {core.material} clad with {core.thickness}µm
- {box.material} clad with {box.thickness}µm"""
)
1
2
3
4
Stack:
- sio2 clad with 3.0µm
- si clad with 0.22µm
- sio2 clad with 3.0µm
2.5D using effective index
Source Code
1
2
3
4
5
6
7
8
9
10
core_material = gplugins.get_effective_indices(
core_material=3.4777,
clad_materialding=1.444,
nsubstrate=1.444,
thickness=0.22,
wavelength=1.55,
polarization="te",
)[0]
core_material
1
2.8494636999424405
Source Code
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
# Define materials
Si = mp.Medium(index=3.45)
SiO2 = mp.Medium(index=1.45)
resolution = 40
dpml = 1
pad = 1
s = gm.write_sparameters_meep_mpi(
opt_device,
cores=16,
xmargin_left=1,
xmargin_right=1,
ymargin_top=1,
ymargin_bot=1,
port_source_names=['o1', 'o2', 'o3'],
port_source_modes={'o1':[0], 'o2':[0], 'o3':[0]},
port_modes=[0],
filepath=Path(f'/home/ramprakash/Integrated_Tests/test_outputs/opt_design_new_2.npz'),
tpml=dpml,
# extend_ports_length=0, # Extend ports to create space for sources/monitors
resolution=resolution,
wavelength_start=wavelengths[0],
wavelength_stop=wavelengths[-1],
wavelength_points=len(wavelengths),
port_source_offset=-0.5,
port_monitor_offset=-0.1,
distance_source_to_monitors=0.3,
# port_symmetries=port_symmetries_coupler,
layer_stack=layer_stack,
material_name_to_meep=dict(si=core_material),
is_3d=False,
overwrite=True,
run=False
)
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
118
119
120
121
122
123
124
125
[32m2025-12-08 23:26:12.782[0m | [1mINFO [0m | [36mgplugins.gmeep.write_sparameters_meep_mpi[0m:[36mwrite_sparameters_meep_mpi[0m:[36m159[0m - [1mWrite PosixPath('/tmp/gdsfactory/temp/write_sparameters_meep_mpi.json')[0m
[32m2025-12-08 23:26:12.794[0m | [1mINFO [0m | [36mgplugins.gmeep.write_sparameters_meep_mpi[0m:[36mwrite_sparameters_meep_mpi[0m:[36m195[0m - [1mmpirun -np 16 /home/ramprakash/anaconda3/envs/si_photo/bin/python /tmp/gdsfactory/temp/write_sparameters_meep_mpi.py[0m
[32m2025-12-08 23:26:12.795[0m | [1mINFO [0m | [36mgplugins.gmeep.write_sparameters_meep_mpi[0m:[36mwrite_sparameters_meep_mpi[0m:[36m196[0m - [1m/home/ramprakash/Integrated_Tests/test_outputs/opt_design_new_2.npz[0m
Using MPI version 4.1, 16 processes
2025-12-08 23:26:17.044 | INFO | gplugins.gmeep:<module>:39 - Meep '1.31.0' installed at ['/home/ramprakash/anaconda3/envs/si_photo/lib/python3.13/site-packages/meep']
2025-12-08 23:29:38.841 | INFO | gplugins.gmeep.write_sparameters_meep:write_sparameters_meep:583 - Write simulation results to PosixPath('/home/ramprakash/Integrated_Tests/test_outputs/opt_design_new_2.npz')
2025-12-08 23:29:39.211 | INFO | gplugins.gmeep.write_sparameters_meep:write_sparameters_meep:585 - Write simulation settings to PosixPath('/home/ramprakash/Integrated_Tests/test_outputs/opt_design_new_2.yml')
Elapsed run time = 204.0357 s
/tmp/gdsfactory/temp/write_sparameters_meep_mpi.py:14: PydanticDeprecatedSince20: The `parse_raw` method is deprecated; if your data is JSON use `model_validate_json`, otherwise load the data then use `model_validate` instead. Deprecated in Pydantic V2.0 to be removed in V3.0. See Pydantic V2 Migration Guide at https://errors.pydantic.dev/2.12/migration/
layer_stack = LayerStack.parse_raw(filepath_json.read_text())
/tmp/gdsfactory/temp/write_sparameters_meep_mpi.py:14: PydanticDeprecatedSince20: The `parse_raw` method is deprecated; if your data is JSON use `model_validate_json`, otherwise load the data then use `model_validate` instead. Deprecated in Pydantic V2.0 to be removed in V3.0. See Pydantic V2 Migration Guide at https://errors.pydantic.dev/2.12/migration/
layer_stack = LayerStack.parse_raw(filepath_json.read_text())
/tmp/gdsfactory/temp/write_sparameters_meep_mpi.py:14: PydanticDeprecatedSince20: The `parse_raw` method is deprecated; if your data is JSON use `model_validate_json`, otherwise load the data then use `model_validate` instead. Deprecated in Pydantic V2.0 to be removed in V3.0. See Pydantic V2 Migration Guide at https://errors.pydantic.dev/2.12/migration/
layer_stack = LayerStack.parse_raw(filepath_json.read_text())
/tmp/gdsfactory/temp/write_sparameters_meep_mpi.py:14: PydanticDeprecatedSince20: The `parse_raw` method is deprecated; if your data is JSON use `model_validate_json`, otherwise load the data then use `model_validate` instead. Deprecated in Pydantic V2.0 to be removed in V3.0. See Pydantic V2 Migration Guide at https://errors.pydantic.dev/2.12/migration/
layer_stack = LayerStack.parse_raw(filepath_json.read_text())
/tmp/gdsfactory/temp/write_sparameters_meep_mpi.py:14: PydanticDeprecatedSince20: The `parse_raw` method is deprecated; if your data is JSON use `model_validate_json`, otherwise load the data then use `model_validate` instead. Deprecated in Pydantic V2.0 to be removed in V3.0. See Pydantic V2 Migration Guide at https://errors.pydantic.dev/2.12/migration/
layer_stack = LayerStack.parse_raw(filepath_json.read_text())
0%| | 0/3 [00:00<?, ?it/s]/tmp/gdsfactory/temp/write_sparameters_meep_mpi.py:14: PydanticDeprecatedSince20: The `parse_raw` method is deprecated; if your data is JSON use `model_validate_json`, otherwise load the data then use `model_validate` instead. Deprecated in Pydantic V2.0 to be removed in V3.0. See Pydantic V2 Migration Guide at https://errors.pydantic.dev/2.12/migration/
layer_stack = LayerStack.parse_raw(filepath_json.read_text())
/home/ramprakash/anaconda3/envs/si_photo/lib/python3.13/site-packages/meep/__init__.py:4440: ComplexWarning: Casting complex values to real discards the imaginary part
return _meep.create_structure(cell_size, dft_data_list_, pml_1d_vols_, pml_2d_vols_, pml_3d_vols_, absorber_vols_, gv, br, sym, num_chunks, Courant, use_anisotropic_averaging, tol, maxeval, gobj_list, center, _ensure_periodicity, _default_material, alist, extra_materials, split_chunks_evenly, set_materials, existing_s, output_chunk_costs, my_bp)
0%| | 0/3 [00:00<?, ?it/s]/tmp/gdsfactory/temp/write_sparameters_meep_mpi.py:14: PydanticDeprecatedSince20: The `parse_raw` method is deprecated; if your data is JSON use `model_validate_json`, otherwise load the data then use `model_validate` instead. Deprecated in Pydantic V2.0 to be removed in V3.0. See Pydantic V2 Migration Guide at https://errors.pydantic.dev/2.12/migration/
layer_stack = LayerStack.parse_raw(filepath_json.read_text())
/tmp/gdsfactory/temp/write_sparameters_meep_mpi.py:14: PydanticDeprecatedSince20: The `parse_raw` method is deprecated; if your data is JSON use `model_validate_json`, otherwise load the data then use `model_validate` instead. Deprecated in Pydantic V2.0 to be removed in V3.0. See Pydantic V2 Migration Guide at https://errors.pydantic.dev/2.12/migration/
layer_stack = LayerStack.parse_raw(filepath_json.read_text())
/tmp/gdsfactory/temp/write_sparameters_meep_mpi.py:14: PydanticDeprecatedSince20: The `parse_raw` method is deprecated; if your data is JSON use `model_validate_json`, otherwise load the data then use `model_validate` instead. Deprecated in Pydantic V2.0 to be removed in V3.0. See Pydantic V2 Migration Guide at https://errors.pydantic.dev/2.12/migration/
layer_stack = LayerStack.parse_raw(filepath_json.read_text())
/tmp/gdsfactory/temp/write_sparameters_meep_mpi.py:14: PydanticDeprecatedSince20: The `parse_raw` method is deprecated; if your data is JSON use `model_validate_json`, otherwise load the data then use `model_validate` instead. Deprecated in Pydantic V2.0 to be removed in V3.0. See Pydantic V2 Migration Guide at https://errors.pydantic.dev/2.12/migration/
layer_stack = LayerStack.parse_raw(filepath_json.read_text())
/home/ramprakash/anaconda3/envs/si_photo/lib/python3.13/site-packages/meep/__init__.py:4440: ComplexWarning: Casting complex values to real discards the imaginary part
return _meep.create_structure(cell_size, dft_data_list_, pml_1d_vols_, pml_2d_vols_, pml_3d_vols_, absorber_vols_, gv, br, sym, num_chunks, Courant, use_anisotropic_averaging, tol, maxeval, gobj_list, center, _ensure_periodicity, _default_material, alist, extra_materials, split_chunks_evenly, set_materials, existing_s, output_chunk_costs, my_bp)
/home/ramprakash/anaconda3/envs/si_photo/lib/python3.13/site-packages/meep/__init__.py:4440: ComplexWarning: Casting complex values to real discards the imaginary part
return _meep.create_structure(cell_size, dft_data_list_, pml_1d_vols_, pml_2d_vols_, pml_3d_vols_, absorber_vols_, gv, br, sym, num_chunks, Courant, use_anisotropic_averaging, tol, maxeval, gobj_list, center, _ensure_periodicity, _default_material, alist, extra_materials, split_chunks_evenly, set_materials, existing_s, output_chunk_costs, my_bp)
0%| | 0/3 [00:00<?, ?it/s]/tmp/gdsfactory/temp/write_sparameters_meep_mpi.py:14: PydanticDeprecatedSince20: The `parse_raw` method is deprecated; if your data is JSON use `model_validate_json`, otherwise load the data then use `model_validate` instead. Deprecated in Pydantic V2.0 to be removed in V3.0. See Pydantic V2 Migration Guide at https://errors.pydantic.dev/2.12/migration/
layer_stack = LayerStack.parse_raw(filepath_json.read_text())
/home/ramprakash/anaconda3/envs/si_photo/lib/python3.13/site-packages/meep/__init__.py:4440: ComplexWarning: Casting complex values to real discards the imaginary part
return _meep.create_structure(cell_size, dft_data_list_, pml_1d_vols_, pml_2d_vols_, pml_3d_vols_, absorber_vols_, gv, br, sym, num_chunks, Courant, use_anisotropic_averaging, tol, maxeval, gobj_list, center, _ensure_periodicity, _default_material, alist, extra_materials, split_chunks_evenly, set_materials, existing_s, output_chunk_costs, my_bp)
/tmp/gdsfactory/temp/write_sparameters_meep_mpi.py:14: PydanticDeprecatedSince20: The `parse_raw` method is deprecated; if your data is JSON use `model_validate_json`, otherwise load the data then use `model_validate` instead. Deprecated in Pydantic V2.0 to be removed in V3.0. See Pydantic V2 Migration Guide at https://errors.pydantic.dev/2.12/migration/
layer_stack = LayerStack.parse_raw(filepath_json.read_text())
/tmp/gdsfactory/temp/write_sparameters_meep_mpi.py:14: PydanticDeprecatedSince20: The `parse_raw` method is deprecated; if your data is JSON use `model_validate_json`, otherwise load the data then use `model_validate` instead. Deprecated in Pydantic V2.0 to be removed in V3.0. See Pydantic V2 Migration Guide at https://errors.pydantic.dev/2.12/migration/
layer_stack = LayerStack.parse_raw(filepath_json.read_text())
/home/ramprakash/anaconda3/envs/si_photo/lib/python3.13/site-packages/meep/__init__.py:4440: ComplexWarning: Casting complex values to real discards the imaginary part
return _meep.create_structure(cell_size, dft_data_list_, pml_1d_vols_, pml_2d_vols_, pml_3d_vols_, absorber_vols_, gv, br, sym, num_chunks, Courant, use_anisotropic_averaging, tol, maxeval, gobj_list, center, _ensure_periodicity, _default_material, alist, extra_materials, split_chunks_evenly, set_materials, existing_s, output_chunk_costs, my_bp)
0%| | 0/3 [00:00<?, ?it/s]/tmp/gdsfactory/temp/write_sparameters_meep_mpi.py:14: PydanticDeprecatedSince20: The `parse_raw` method is deprecated; if your data is JSON use `model_validate_json`, otherwise load the data then use `model_validate` instead. Deprecated in Pydantic V2.0 to be removed in V3.0. See Pydantic V2 Migration Guide at https://errors.pydantic.dev/2.12/migration/
layer_stack = LayerStack.parse_raw(filepath_json.read_text())
0%| | 0/3 [00:00<?, ?it/s]/tmp/gdsfactory/temp/write_sparameters_meep_mpi.py:14: PydanticDeprecatedSince20: The `parse_raw` method is deprecated; if your data is JSON use `model_validate_json`, otherwise load the data then use `model_validate` instead. Deprecated in Pydantic V2.0 to be removed in V3.0. See Pydantic V2 Migration Guide at https://errors.pydantic.dev/2.12/migration/
layer_stack = LayerStack.parse_raw(filepath_json.read_text())
/tmp/gdsfactory/temp/write_sparameters_meep_mpi.py:14: PydanticDeprecatedSince20: The `parse_raw` method is deprecated; if your data is JSON use `model_validate_json`, otherwise load the data then use `model_validate` instead. Deprecated in Pydantic V2.0 to be removed in V3.0. See Pydantic V2 Migration Guide at https://errors.pydantic.dev/2.12/migration/
layer_stack = LayerStack.parse_raw(filepath_json.read_text())
0%| | 0/3 [00:00<?, ?it/s]/home/ramprakash/anaconda3/envs/si_photo/lib/python3.13/site-packages/meep/__init__.py:4440: ComplexWarning: Casting complex values to real discards the imaginary part
return _meep.create_structure(cell_size, dft_data_list_, pml_1d_vols_, pml_2d_vols_, pml_3d_vols_, absorber_vols_, gv, br, sym, num_chunks, Courant, use_anisotropic_averaging, tol, maxeval, gobj_list, center, _ensure_periodicity, _default_material, alist, extra_materials, split_chunks_evenly, set_materials, existing_s, output_chunk_costs, my_bp)
0%| | 0/3 [00:00<?, ?it/s]/home/ramprakash/anaconda3/envs/si_photo/lib/python3.13/site-packages/meep/__init__.py:4440: ComplexWarning: Casting complex values to real discards the imaginary part
return _meep.create_structure(cell_size, dft_data_list_, pml_1d_vols_, pml_2d_vols_, pml_3d_vols_, absorber_vols_, gv, br, sym, num_chunks, Courant, use_anisotropic_averaging, tol, maxeval, gobj_list, center, _ensure_periodicity, _default_material, alist, extra_materials, split_chunks_evenly, set_materials, existing_s, output_chunk_costs, my_bp)
/home/ramprakash/anaconda3/envs/si_photo/lib/python3.13/site-packages/meep/__init__.py:4440: ComplexWarning: Casting complex values to real discards the imaginary part
return _meep.create_structure(cell_size, dft_data_list_, pml_1d_vols_, pml_2d_vols_, pml_3d_vols_, absorber_vols_, gv, br, sym, num_chunks, Courant, use_anisotropic_averaging, tol, maxeval, gobj_list, center, _ensure_periodicity, _default_material, alist, extra_materials, split_chunks_evenly, set_materials, existing_s, output_chunk_costs, my_bp)
/home/ramprakash/anaconda3/envs/si_photo/lib/python3.13/site-packages/meep/__init__.py:4440: ComplexWarning: Casting complex values to real discards the imaginary part
return _meep.create_structure(cell_size, dft_data_list_, pml_1d_vols_, pml_2d_vols_, pml_3d_vols_, absorber_vols_, gv, br, sym, num_chunks, Courant, use_anisotropic_averaging, tol, maxeval, gobj_list, center, _ensure_periodicity, _default_material, alist, extra_materials, split_chunks_evenly, set_materials, existing_s, output_chunk_costs, my_bp)
/home/ramprakash/anaconda3/envs/si_photo/lib/python3.13/site-packages/meep/__init__.py:4440: ComplexWarning: Casting complex values to real discards the imaginary part
return _meep.create_structure(cell_size, dft_data_list_, pml_1d_vols_, pml_2d_vols_, pml_3d_vols_, absorber_vols_, gv, br, sym, num_chunks, Courant, use_anisotropic_averaging, tol, maxeval, gobj_list, center, _ensure_periodicity, _default_material, alist, extra_materials, split_chunks_evenly, set_materials, existing_s, output_chunk_costs, my_bp)
0%| | 0/3 [00:00<?, ?it/s]/home/ramprakash/anaconda3/envs/si_photo/lib/python3.13/site-packages/meep/__init__.py:4440: ComplexWarning: Casting complex values to real discards the imaginary part
return _meep.create_structure(cell_size, dft_data_list_, pml_1d_vols_, pml_2d_vols_, pml_3d_vols_, absorber_vols_, gv, br, sym, num_chunks, Courant, use_anisotropic_averaging, tol, maxeval, gobj_list, center, _ensure_periodicity, _default_material, alist, extra_materials, split_chunks_evenly, set_materials, existing_s, output_chunk_costs, my_bp)
0%| | 0/3 [00:00<?, ?it/s]/home/ramprakash/anaconda3/envs/si_photo/lib/python3.13/site-packages/meep/__init__.py:4440: ComplexWarning: Casting complex values to real discards the imaginary part
return _meep.create_structure(cell_size, dft_data_list_, pml_1d_vols_, pml_2d_vols_, pml_3d_vols_, absorber_vols_, gv, br, sym, num_chunks, Courant, use_anisotropic_averaging, tol, maxeval, gobj_list, center, _ensure_periodicity, _default_material, alist, extra_materials, split_chunks_evenly, set_materials, existing_s, output_chunk_costs, my_bp)
0%| | 0/3 [00:00<?, ?it/s]/home/ramprakash/anaconda3/envs/si_photo/lib/python3.13/site-packages/meep/__init__.py:4440: ComplexWarning: Casting complex values to real discards the imaginary part
return _meep.create_structure(cell_size, dft_data_list_, pml_1d_vols_, pml_2d_vols_, pml_3d_vols_, absorber_vols_, gv, br, sym, num_chunks, Courant, use_anisotropic_averaging, tol, maxeval, gobj_list, center, _ensure_periodicity, _default_material, alist, extra_materials, split_chunks_evenly, set_materials, existing_s, output_chunk_costs, my_bp)
Warning: grid volume is not an integer number of pixels; cell size will be rounded to nearest pixel.
/home/ramprakash/anaconda3/envs/si_photo/lib/python3.13/site-packages/meep/__init__.py:4440: ComplexWarning: Casting complex values to real discards the imaginary part
return _meep.create_structure(cell_size, dft_data_list_, pml_1d_vols_, pml_2d_vols_, pml_3d_vols_, absorber_vols_, gv, br, sym, num_chunks, Courant, use_anisotropic_averaging, tol, maxeval, gobj_list, center, _ensure_periodicity, _default_material, alist, extra_materials, split_chunks_evenly, set_materials, existing_s, output_chunk_costs, my_bp)
/home/ramprakash/anaconda3/envs/si_photo/lib/python3.13/site-packages/meep/__init__.py:4440: ComplexWarning: Casting complex values to real discards the imaginary part
return _meep.create_structure(cell_size, dft_data_list_, pml_1d_vols_, pml_2d_vols_, pml_3d_vols_, absorber_vols_, gv, br, sym, num_chunks, Courant, use_anisotropic_averaging, tol, maxeval, gobj_list, center, _ensure_periodicity, _default_material, alist, extra_materials, split_chunks_evenly, set_materials, existing_s, output_chunk_costs, my_bp)
/home/ramprakash/anaconda3/envs/si_photo/lib/python3.13/site-packages/meep/__init__.py:4440: ComplexWarning: Casting complex values to real discards the imaginary part
return _meep.create_structure(cell_size, dft_data_list_, pml_1d_vols_, pml_2d_vols_, pml_3d_vols_, absorber_vols_, gv, br, sym, num_chunks, Courant, use_anisotropic_averaging, tol, maxeval, gobj_list, center, _ensure_periodicity, _default_material, alist, extra_materials, split_chunks_evenly, set_materials, existing_s, output_chunk_costs, my_bp)
/home/ramprakash/anaconda3/envs/si_photo/lib/python3.13/site-packages/meep/__init__.py:4443: ComplexWarning: Casting complex values to real discards the imaginary part
return _meep._set_materials(s, cell_size, gv, use_anisotropic_averaging, tol, maxeval, gobj_list, center, _ensure_periodicity, _default_material, alist, extra_materials, split_chunks_evenly, set_materials, existing_geps, output_chunk_costs, my_bp)
/home/ramprakash/anaconda3/envs/si_photo/lib/python3.13/site-packages/meep/__init__.py:4443: ComplexWarning: Casting complex values to real discards the imaginary part
return _meep._set_materials(s, cell_size, gv, use_anisotropic_averaging, tol, maxeval, gobj_list, center, _ensure_periodicity, _default_material, alist, extra_materials, split_chunks_evenly, set_materials, existing_geps, output_chunk_costs, my_bp)
/home/ramprakash/anaconda3/envs/si_photo/lib/python3.13/site-packages/meep/__init__.py:4443: ComplexWarning: Casting complex values to real discards the imaginary part
return _meep._set_materials(s, cell_size, gv, use_anisotropic_averaging, tol, maxeval, gobj_list, center, _ensure_periodicity, _default_material, alist, extra_materials, split_chunks_evenly, set_materials, existing_geps, output_chunk_costs, my_bp)
/home/ramprakash/anaconda3/envs/si_photo/lib/python3.13/site-packages/meep/__init__.py:4443: ComplexWarning: Casting complex values to real discards the imaginary part
return _meep._set_materials(s, cell_size, gv, use_anisotropic_averaging, tol, maxeval, gobj_list, center, _ensure_periodicity, _default_material, alist, extra_materials, split_chunks_evenly, set_materials, existing_geps, output_chunk_costs, my_bp)
/home/ramprakash/anaconda3/envs/si_photo/lib/python3.13/site-packages/meep/__init__.py:4443: ComplexWarning: Casting complex values to real discards the imaginary part
return _meep._set_materials(s, cell_size, gv, use_anisotropic_averaging, tol, maxeval, gobj_list, center, _ensure_periodicity, _default_material, alist, extra_materials, split_chunks_evenly, set_materials, existing_geps, output_chunk_costs, my_bp)
/home/ramprakash/anaconda3/envs/si_photo/lib/python3.13/site-packages/meep/__init__.py:4443: ComplexWarning: Casting complex values to real discards the imaginary part
return _meep._set_materials(s, cell_size, gv, use_anisotropic_averaging, tol, maxeval, gobj_list, center, _ensure_periodicity, _default_material, alist, extra_materials, split_chunks_evenly, set_materials, existing_geps, output_chunk_costs, my_bp)
/home/ramprakash/anaconda3/envs/si_photo/lib/python3.13/site-packages/meep/__init__.py:4443: ComplexWarning: Casting complex values to real discards the imaginary part
return _meep._set_materials(s, cell_size, gv, use_anisotropic_averaging, tol, maxeval, gobj_list, center, _ensure_periodicity, _default_material, alist, extra_materials, split_chunks_evenly, set_materials, existing_geps, output_chunk_costs, my_bp)
/home/ramprakash/anaconda3/envs/si_photo/lib/python3.13/site-packages/meep/__init__.py:4443: ComplexWarning: Casting complex values to real discards the imaginary part
return _meep._set_materials(s, cell_size, gv, use_anisotropic_averaging, tol, maxeval, gobj_list, center, _ensure_periodicity, _default_material, alist, extra_materials, split_chunks_evenly, set_materials, existing_geps, output_chunk_costs, my_bp)
/home/ramprakash/anaconda3/envs/si_photo/lib/python3.13/site-packages/meep/__init__.py:4443: ComplexWarning: Casting complex values to real discards the imaginary part
return _meep._set_materials(s, cell_size, gv, use_anisotropic_averaging, tol, maxeval, gobj_list, center, _ensure_periodicity, _default_material, alist, extra_materials, split_chunks_evenly, set_materials, existing_geps, output_chunk_costs, my_bp)
/home/ramprakash/anaconda3/envs/si_photo/lib/python3.13/site-packages/meep/__init__.py:4443: ComplexWarning: Casting complex values to real discards the imaginary part
return _meep._set_materials(s, cell_size, gv, use_anisotropic_averaging, tol, maxeval, gobj_list, center, _ensure_periodicity, _default_material, alist, extra_materials, split_chunks_evenly, set_materials, existing_geps, output_chunk_costs, my_bp)
/home/ramprakash/anaconda3/envs/si_photo/lib/python3.13/site-packages/meep/__init__.py:4443: ComplexWarning: Casting complex values to real discards the imaginary part
return _meep._set_materials(s, cell_size, gv, use_anisotropic_averaging, tol, maxeval, gobj_list, center, _ensure_periodicity, _default_material, alist, extra_materials, split_chunks_evenly, set_materials, existing_geps, output_chunk_costs, my_bp)
/home/ramprakash/anaconda3/envs/si_photo/lib/python3.13/site-packages/meep/__init__.py:4443: ComplexWarning: Casting complex values to real discards the imaginary part
return _meep._set_materials(s, cell_size, gv, use_anisotropic_averaging, tol, maxeval, gobj_list, center, _ensure_periodicity, _default_material, alist, extra_materials, split_chunks_evenly, set_materials, existing_geps, output_chunk_costs, my_bp)
/home/ramprakash/anaconda3/envs/si_photo/lib/python3.13/site-packages/meep/__init__.py:4443: ComplexWarning: Casting complex values to real discards the imaginary part
return _meep._set_materials(s, cell_size, gv, use_anisotropic_averaging, tol, maxeval, gobj_list, center, _ensure_periodicity, _default_material, alist, extra_materials, split_chunks_evenly, set_materials, existing_geps, output_chunk_costs, my_bp)
/home/ramprakash/anaconda3/envs/si_photo/lib/python3.13/site-packages/meep/__init__.py:4443: ComplexWarning: Casting complex values to real discards the imaginary part
return _meep._set_materials(s, cell_size, gv, use_anisotropic_averaging, tol, maxeval, gobj_list, center, _ensure_periodicity, _default_material, alist, extra_materials, split_chunks_evenly, set_materials, existing_geps, output_chunk_costs, my_bp)
/home/ramprakash/anaconda3/envs/si_photo/lib/python3.13/site-packages/meep/__init__.py:4443: ComplexWarning: Casting complex values to real discards the imaginary part
return _meep._set_materials(s, cell_size, gv, use_anisotropic_averaging, tol, maxeval, gobj_list, center, _ensure_periodicity, _default_material, alist, extra_materials, split_chunks_evenly, set_materials, existing_geps, output_chunk_costs, my_bp)
/home/ramprakash/anaconda3/envs/si_photo/lib/python3.13/site-packages/meep/__init__.py:4443: ComplexWarning: Casting complex values to real discards the imaginary part
return _meep._set_materials(s, cell_size, gv, use_anisotropic_averaging, tol, maxeval, gobj_list, center, _ensure_periodicity, _default_material, alist, extra_materials, split_chunks_evenly, set_materials, existing_geps, output_chunk_costs, my_bp)
33%|██████████████▋ | 1/3 [00:37<01:15, 37.61s/it]Warning: grid volume is not an integer number of pixels; cell size will be rounded to nearest pixel.
67%|█████████████████████████████▎ | 2/3 [01:59<01:03, 63.56s/it]Warning: grid volume is not an integer number of pixels; cell size will be rounded to nearest pixel.
100%|████████████████████████████████████████████| 3/3 [03:21<00:00, 67.26s/it]
100%|████████████████████████████████████████████| 3/3 [03:21<00:00, 67.26s/it]
100%|████████████████████████████████████████████| 3/3 [03:21<00:00, 67.29s/it]
100%|████████████████████████████████████████████| 3/3 [03:21<00:00, 67.26s/it]
100%|████████████████████████████████████████████| 3/3 [03:21<00:00, 67.24s/it]
100%|████████████████████████████████████████████| 3/3 [03:21<00:00, 67.29s/it]
100%|████████████████████████████████████████████| 3/3 [03:21<00:00, 67.24s/it]
100%|████████████████████████████████████████████| 3/3 [03:21<00:00, 67.24s/it]
100%|████████████████████████████████████████████| 3/3 [03:21<00:00, 67.26s/it]
100%|████████████████████████████████████████████| 3/3 [03:21<00:00, 67.28s/it]
100%|████████████████████████████████████████████| 3/3 [03:21<00:00, 67.29s/it]
100%|████████████████████████████████████████████| 3/3 [03:21<00:00, 67.26s/it]
100%|████████████████████████████████████████████| 3/3 [03:21<00:00, 67.25s/it]
100%|████████████████████████████████████████████| 3/3 [03:21<00:00, 67.25s/it]
100%|████████████████████████████████████████████| 3/3 [03:21<00:00, 67.26s/it]
Source Code
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
# Calculate insertion loss from S-parameters
sp = np.load(s)
# Extract S-parameters at center wavelength
wavelengths_sim = sp['wavelengths']
center_idx = len(wavelengths_sim) // 2
# S21 and S31 (transmission from port 1 to ports 2 and 3)
S21 = sp['o1@0,o2@0'][center_idx] # Port 1 to Port 2 (top)
S31 = sp['o1@0,o3@0'][center_idx] # Port 1 to Port 3 (bottom)
S11 = sp['o1@0,o1@0'][center_idx] # Reflection at Port 1
# Calculate power transmission (magnitude squared)
T21 = np.abs(S21)**2 # Transmission to port 2
T31 = np.abs(S31)**2 # Transmission to port 3
R11 = np.abs(S11)**2 # Reflection
# Total transmission
total_transmission = T21 + T31
# Insertion loss (in dB)
insertion_loss_dB = -10 * np.log10(total_transmission)
# Individual port losses
S21_dB = 10 * np.log10(T21)
S31_dB = 10 * np.log10(T31)
S11_dB = 10 * np.log10(R11)
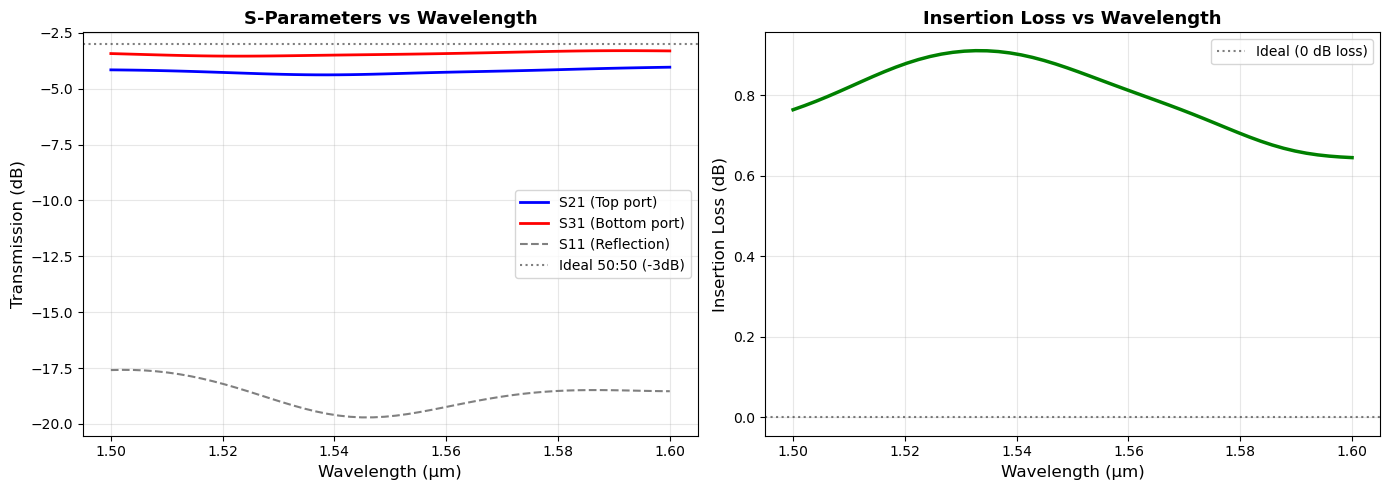
print("="*70)
print("S-PARAMETER ANALYSIS (at λ = {:.3f} μm)".format(wavelengths_sim[center_idx]))
print("="*70)
print("\nTransmission Coefficients:")
print(f" S21 (to top port): {S21_dB:.2f} dB ({T21:.4f} or {T21*100:.2f}%)")
print(f" S31 (to bottom port): {S31_dB:.2f} dB ({T31:.4f} or {T31*100:.2f}%)")
print(f"\nReflection:")
print(f" S11 (reflection): {S11_dB:.2f} dB ({R11:.4f} or {R11*100:.2f}%)")
print(f"\nTotal Performance:")
print(f" Total Transmission: {total_transmission:.4f} ({total_transmission*100:.2f}%)")
print(f" Insertion Loss: {insertion_loss_dB:.2f} dB")
print(f"\nPower Balance Check:")
print(f" Transmitted + Reflected: {(total_transmission + R11)*100:.2f}%")
print(f" Lost (scattering/absorption): {(1 - total_transmission - R11)*100:.2f}%")
print(f"\nSplitting Performance:")
imbalance = abs(S21_dB - S31_dB)
print(f" Imbalance: {imbalance:.2f} dB")
if imbalance < 0.5:
print(f" Quality: Excellent (<0.5 dB)")
elif imbalance < 1.0:
print(f" Quality: Good (<1 dB)")
else:
print(f" Quality: Fair (>{imbalance:.1f} dB)")
print("="*70)
# Plot S-parameters vs wavelength
fig, axes = plt.subplots(1, 2, figsize=(14, 5))
# Plot 1: Transmission in dB
ax = axes[0]
S21_dB_all = 10 * np.log10(np.abs(sp['o1@0,o2@0'])**2)
S31_dB_all = 10 * np.log10(np.abs(sp['o1@0,o3@0'])**2)
S11_dB_all = 10 * np.log10(np.abs(sp['o1@0,o1@0'])**2)
ax.plot(wavelengths_sim, S21_dB_all, 'b-', linewidth=2, label='S21 (Top port)')
ax.plot(wavelengths_sim, S31_dB_all, 'r-', linewidth=2, label='S31 (Bottom port)')
ax.plot(wavelengths_sim, S11_dB_all, 'gray', linewidth=1.5, linestyle='--', label='S11 (Reflection)')
ax.axhline(y=-3, color='k', linestyle=':', alpha=0.5, label='Ideal 50:50 (-3dB)')
ax.set_xlabel('Wavelength (μm)', fontsize=12)
ax.set_ylabel('Transmission (dB)', fontsize=12)
ax.set_title('S-Parameters vs Wavelength', fontsize=13, fontweight='bold')
ax.legend()
ax.grid(True, alpha=0.3)
# Plot 2: Insertion Loss
ax = axes[1]
total_trans_all = np.abs(sp['o1@0,o2@0'])**2 + np.abs(sp['o1@0,o3@0'])**2
insertion_loss_all = -10 * np.log10(total_trans_all)
ax.plot(wavelengths_sim, insertion_loss_all, 'g-', linewidth=2.5)
ax.axhline(y=0, color='k', linestyle=':', alpha=0.5, label='Ideal (0 dB loss)')
ax.set_xlabel('Wavelength (μm)', fontsize=12)
ax.set_ylabel('Insertion Loss (dB)', fontsize=12)
ax.set_title('Insertion Loss vs Wavelength', fontsize=13, fontweight='bold')
ax.legend()
ax.grid(True, alpha=0.3)
plt.tight_layout()
plt.savefig('insertion_loss_analysis.png', dpi=150, bbox_inches='tight')
plt.show()
# print("\nPlot saved as 'insertion_loss_analysis.png'")
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
======================================================================
S-PARAMETER ANALYSIS (at λ = 1.551 μm)
======================================================================
Transmission Coefficients:
S21 (to top port): -4.32 dB (0.3697 or 36.97%)
S31 (to bottom port): -3.46 dB (0.4509 or 45.09%)
Reflection:
S11 (reflection): -19.63 dB (0.0109 or 1.09%)
Total Performance:
Total Transmission: 0.8206 (82.06%)
Insertion Loss: 0.86 dB
Power Balance Check:
Transmitted + Reflected: 83.14%
Lost (scattering/absorption): 16.86%
Splitting Performance:
Imbalance: 0.86 dB
Quality: Good (<1 dB)
======================================================================
For generic device from gdsfactory
Source Code
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
# Define materials
Si = mp.Medium(index=3.45)
SiO2 = mp.Medium(index=1.45)
resolution = 20
dpml = 1
pad = 1
s_ybranch = gm.write_sparameters_meep_mpi(
ybranch,
cores=8,
xmargin_left=1,
xmargin_right=1,
ymargin_top=1,
ymargin_bot=1,
port_source_names=['o1', 'o2', 'o3'],
port_source_modes={'o1':[0], 'o2':[0], 'o3':[0]},
port_modes=[0],
filepath=Path(f'/home/ramprakash/Integrated_Tests/test_outputs/y_branch_sparameters.npz'),
tpml=dpml,
resolution=resolution,
wavelength_start=wavelengths[0],
wavelength_stop=wavelengths[-1],
wavelength_points=len(wavelengths),
port_source_offset=-0.5,
port_monitor_offset=-0.1,
distance_source_to_monitors=0.3,
# port_symmetries=port_symmetries_coupler,
layer_stack=layer_stack,
material_name_to_meep=dict(si=core_material),
is_3d=False,
overwrite=False,
run=False
)
1
[32m2025-12-08 23:30:54.555[0m | [1mINFO [0m | [36mgplugins.gmeep.write_sparameters_meep_mpi[0m:[36mwrite_sparameters_meep_mpi[0m:[36m148[0m - [1mSimulation PosixPath('/home/ramprakash/Integrated_Tests/test_outputs/y_branch_sparameters.npz') already exists[0m
Source Code
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
# Calculate insertion loss from S-parameters
sp = np.load(s_ybranch)
# Extract S-parameters at center wavelength
wavelengths_sim = sp['wavelengths']
center_idx = len(wavelengths_sim) // 2
# S21 and S31 (transmission from port 1 to ports 2 and 3)
S21 = sp['o1@0,o2@0'][center_idx] # Port 1 to Port 2 (top)
S31 = sp['o1@0,o3@0'][center_idx] # Port 1 to Port 3 (bottom)
S11 = sp['o1@0,o1@0'][center_idx] # Reflection at Port 1
# Calculate power transmission (magnitude squared)
T21 = np.abs(S21)**2 # Transmission to port 2
T31 = np.abs(S31)**2 # Transmission to port 3
R11 = np.abs(S11)**2 # Reflection
# Total transmission
total_transmission = T21 + T31
# Insertion loss (in dB)
insertion_loss_dB = -10 * np.log10(total_transmission)
# Individual port losses
S21_dB = 10 * np.log10(T21)
S31_dB = 10 * np.log10(T31)
S11_dB = 10 * np.log10(R11)
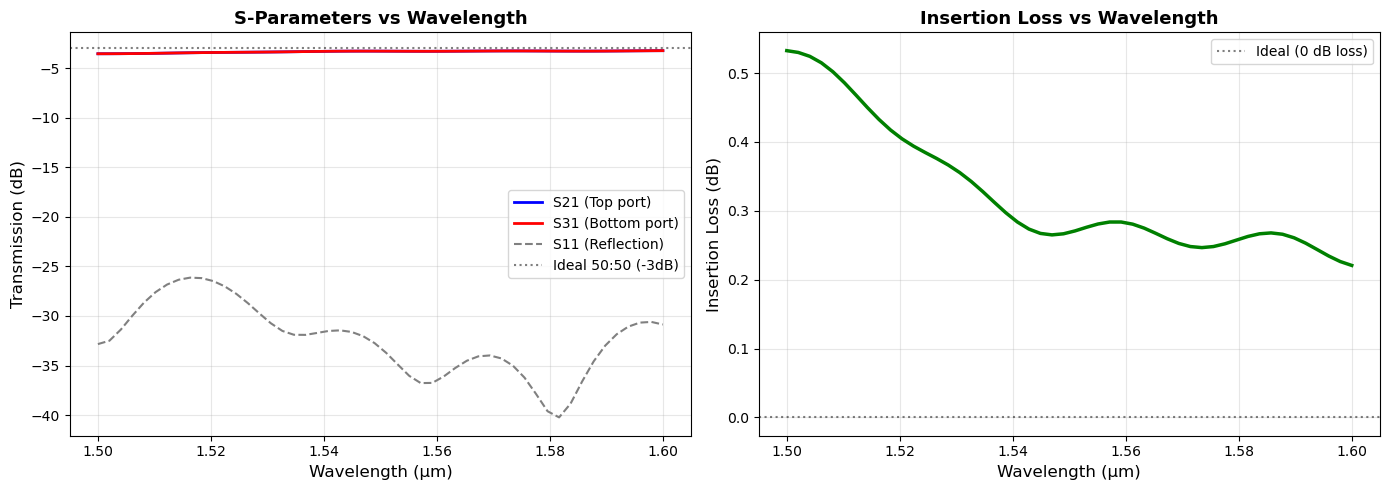
print("="*70)
print("S-PARAMETER ANALYSIS (at λ = {:.3f} μm)".format(wavelengths_sim[center_idx]))
print("="*70)
print("\nTransmission Coefficients:")
print(f" S21 (to top port): {S21_dB:.2f} dB ({T21:.4f} or {T21*100:.2f}%)")
print(f" S31 (to bottom port): {S31_dB:.2f} dB ({T31:.4f} or {T31*100:.2f}%)")
print(f"\nReflection:")
print(f" S11 (reflection): {S11_dB:.2f} dB ({R11:.4f} or {R11*100:.2f}%)")
print(f"\nTotal Performance:")
print(f" Total Transmission: {total_transmission:.4f} ({total_transmission*100:.2f}%)")
print(f" Insertion Loss: {insertion_loss_dB:.2f} dB")
print(f"\nPower Balance Check:")
print(f" Transmitted + Reflected: {(total_transmission + R11)*100:.2f}%")
print(f" Lost (scattering/absorption): {(1 - total_transmission - R11)*100:.2f}%")
print(f"\nSplitting Performance:")
imbalance = abs(S21_dB - S31_dB)
print(f" Imbalance: {imbalance:.2f} dB")
if imbalance < 0.5:
print(f" Quality: Excellent (<0.5 dB)")
elif imbalance < 1.0:
print(f" Quality: Good (<1 dB)")
else:
print(f" Quality: Fair (>{imbalance:.1f} dB)")
print("="*70)
# Plot S-parameters vs wavelength
fig, axes = plt.subplots(1, 2, figsize=(14, 5))
# Plot 1: Transmission in dB
ax = axes[0]
S21_dB_all = 10 * np.log10(np.abs(sp['o1@0,o2@0'])**2)
S31_dB_all = 10 * np.log10(np.abs(sp['o1@0,o3@0'])**2)
S11_dB_all = 10 * np.log10(np.abs(sp['o1@0,o1@0'])**2)
ax.plot(wavelengths_sim, S21_dB_all, 'b-', linewidth=2, label='S21 (Top port)')
ax.plot(wavelengths_sim, S31_dB_all, 'r-', linewidth=2, label='S31 (Bottom port)')
ax.plot(wavelengths_sim, S11_dB_all, 'gray', linewidth=1.5, linestyle='--', label='S11 (Reflection)')
ax.axhline(y=-3, color='k', linestyle=':', alpha=0.5, label='Ideal 50:50 (-3dB)')
ax.set_xlabel('Wavelength (μm)', fontsize=12)
ax.set_ylabel('Transmission (dB)', fontsize=12)
ax.set_title('S-Parameters vs Wavelength', fontsize=13, fontweight='bold')
ax.legend()
ax.grid(True, alpha=0.3)
# Plot 2: Insertion Loss
ax = axes[1]
total_trans_all = np.abs(sp['o1@0,o2@0'])**2 + np.abs(sp['o1@0,o3@0'])**2
insertion_loss_all = -10 * np.log10(total_trans_all)
ax.plot(wavelengths_sim, insertion_loss_all, 'g-', linewidth=2.5)
ax.axhline(y=0, color='k', linestyle=':', alpha=0.5, label='Ideal (0 dB loss)')
ax.set_xlabel('Wavelength (μm)', fontsize=12)
ax.set_ylabel('Insertion Loss (dB)', fontsize=12)
ax.set_title('Insertion Loss vs Wavelength', fontsize=13, fontweight='bold')
ax.legend()
ax.grid(True, alpha=0.3)
plt.tight_layout()
plt.savefig('insertion_loss_analysis.png', dpi=150, bbox_inches='tight')
plt.show()
# print("\nPlot saved as 'insertion_loss_analysis.png'")
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
======================================================================
S-PARAMETER ANALYSIS (at λ = 1.551 μm)
======================================================================
Transmission Coefficients:
S21 (to top port): -3.28 dB (0.4698 or 46.98%)
S31 (to bottom port): -3.28 dB (0.4698 or 46.98%)
Reflection:
S11 (reflection): -33.72 dB (0.0004 or 0.04%)
Total Performance:
Total Transmission: 0.9395 (93.95%)
Insertion Loss: 0.27 dB
Power Balance Check:
Transmitted + Reflected: 94.00%
Lost (scattering/absorption): 6.00%
Splitting Performance:
Imbalance: 0.00 dB
Quality: Excellent (<0.5 dB)
======================================================================
This post is licensed under CC BY 4.0 by the author.